








Bruce Randall Donald
-- Publications
Professor of Computer
Science, Biochemistry, and Chemistry
Duke University

 Books
Books Molecular
Biology.
Molecular
Biology. 






(in Comm. ACM,
2019).
http://www.youtube.com/user/labdonald/videos, (www.youtube.com/@Donaldlab)
with many videos describing our research.
Most of these videos are on
microrobotics and
nanoscience.
While a lot of our work now is on
structural and computational biology, we have not made many videos
on those topics yet. However, we have
written a textbook and plenty of papers you can read on
structural and computational biology and biochemistry.
However, OspreyTube
is growing.
Please note that not all of our videos above are on YouTube because
some, for example, are hosted
in a lecture series at MIT.
Some of our videos are copyrighted by the music
publisher (because of
the music) and therefore
are here, here,
or here,
or here. Or Here
(Siggraph 1990).
*Indicates indexes and reviews of our papers not written/compiled
by our group.
*Please note: Most indexes and collections of our papers are incomplete since the
computer science indexes do not list the biomedical journals, and
Medline does not list the computer science and robotics papers. Only Google Scholar
|
Predicting Cancer Resistance Mutations by Pareto Optimization Over Multistate Protein Design | ||

PNAS (2010). |

|
|
|
Computational Structure-Based Redesign of Enzyme Activity. PNAS (2009). |
(in Comm. ACM, 2019). |

|
|
Computational design of a tight binder to KRas, an “undruggable” cancer target PLoS Computational Biology 2020. | ||
 |
||
|
Cover Article, Chemistry & Biology 2007. [Caption]
|
||
 |
||
| Redesigning the PheA Domain of Gramicidin Synthetase. Biochemistry 2006; 45 (51):15495-15504. |
||
 |
||
| Structure
of Dihydrofolate Reductase-Thymidylate Synthase from Cryptosporidium
hominis. (J. Biol.
Chem. 2003). |
||
 |
||
| Cover article, J.
Eukaryotic Microbiology (2003). |
||
 |
||
| Cover article, PROTEINS (2012). |
 
Complete ensemble of NMR structures of the unphosphorylated human cardiac phospholamban pentamer. PDB ID: 2HYN (Proteins, 2006) |
|
This journal paper is a revised paper based on the earlier Conference version:
LUTE (Local Unpruned Tuple Expansion): Accurate continuously flexible protein design with general energy functions and rigid-rotamer-like efficiency (with M. Hallen and J. Jou). Proceedings of the Annual International Conference on Research in Computational Molecular Biology (RECOMB), Los Angeles, CA, April, 2016. In Research in Computational Molecular Biology, Lecture Notes in Bioinformatics (LNBI), Springer-Verlag (Berlin), Volume 9649, 2016, pp. 122-136.
Our lab has a YouTube channel: http://www.youtube.com/user/labdonald/videos, with many videos describing our mobile robotics research.
 Distributed
Manipulation of Multiple Objects Using Ropes, (with L. Gariepy and D.
Rus), Proc. IEEE International Conference on Robotics and Automation
(ICRA), San Francisco (April, 2000) pp. 450-457.
Distributed
Manipulation of Multiple Objects Using Ropes, (with L. Gariepy and D.
Rus), Proc. IEEE International Conference on Robotics and Automation
(ICRA), San Francisco (April, 2000) pp. 450-457.
| Movie, 4:30, Quicktime w/Sorenson codec, max 250 kb/s (42MB) |
mapmaking-movie-5.250kbit.mov |
| Movie, 4:30, Quicktime w/Sorenson codec, max 100 kb/s (26MB) |
mapmaking-movie-5.100kbit.mov |
| The paper (Gzipped PostScript) (0.8 MB) | ICRA'2000 paper |

 Visibility-Based
Planning of Sensor Control Strategies, (with Amy Briggs),
Algorithmica 26(3/4), March/April (2000) pp.
364-388.
Visibility-Based
Planning of Sensor Control Strategies, (with Amy Briggs),
Algorithmica 26(3/4), March/April (2000) pp.
364-388.

 Mobile
Robot Self-Localization without Explicit Landmarks, (with R. G. Brown),
Algorithmica 26(3/4), March/April (2000) pp.
515-559.
Mobile
Robot Self-Localization without Explicit Landmarks, (with R. G. Brown),
Algorithmica 26(3/4), March/April (2000) pp.
515-559.
 Experiments in
Constrained Prehensile Manipulation: Distributed Manipulation with Ropes,
with Larry Gariepy and Daniela
Rus, In Experimental Robotics VI, Lecture Notes in Control and Information
Sciences, Vol. 250, eds. P. Corke, Springer-Verlag, (2000).
Experiments in
Constrained Prehensile Manipulation: Distributed Manipulation with Ropes,
with Larry Gariepy and Daniela
Rus, In Experimental Robotics VI, Lecture Notes in Control and Information
Sciences, Vol. 250, eds. P. Corke, Springer-Verlag, (2000).

 Amy Briggs and B. R. Donald, Robust Geometric
Algorithms for Sensor Planning, [TR FTP]
International Workshop on the Algorithmic Foundations of Robotics,
Toulouse, France (1996).
Amy Briggs and B. R. Donald, Robust Geometric
Algorithms for Sensor Planning, [TR FTP]
International Workshop on the Algorithmic Foundations of Robotics,
Toulouse, France (1996).
 Information
Invariants for Distributed Manipulation (with J. Jennings and D. Rus) in International
Journal of Robotics Research, Vol. 16, No. 5 (1997). pp. 673-702.
Information
Invariants for Distributed Manipulation (with J. Jennings and D. Rus) in International
Journal of Robotics Research, Vol. 16, No. 5 (1997). pp. 673-702.
 B. R. Donald, J. Jennings, and D. Rus, Minimalism +
Distribution = Supermodularity [TR FTP], Journal
of Experimental and Theoretical Artificial Intelligence (JETAI), Special
issue on Software Architectures for Hardware Agents; Vol. 9 No. 2-3 (1997) pp.
293-321.
B. R. Donald, J. Jennings, and D. Rus, Minimalism +
Distribution = Supermodularity [TR FTP], Journal
of Experimental and Theoretical Artificial Intelligence (JETAI), Special
issue on Software Architectures for Hardware Agents; Vol. 9 No. 2-3 (1997) pp.
293-321.
 Cost-effective, Capable, and
Configurable Multiple Robots: A Report to the ARPA ISAT Study
Group, Summer, 1996 (with J. Jennings and D. Rus).
Cost-effective, Capable, and
Configurable Multiple Robots: A Report to the ARPA ISAT Study
Group, Summer, 1996 (with J. Jennings and D. Rus).
 Moving Furniture with
Teams of Automonous Mobile Robots [TR FTP], (with J.
Jennings and D. Rus) in
Moving Furniture with
Teams of Automonous Mobile Robots [TR FTP], (with J.
Jennings and D. Rus) in
Proc. IEEE/Robotics Society of Japan International Workshop on Intelligent Robots and Systems, (IROS) Pittsburgh, PA (1995).


 Information
Invariants for Distributed Manipulation [TR
FTP] (with J. Jennings and D.
Rus) in The First Workshop on the Algorithmic Foundations of
Robotics, A. K. Peters, Boston, MA. ed. R. Wilson and J.-C.Latombe (1994).
Information
Invariants for Distributed Manipulation [TR
FTP] (with J. Jennings and D.
Rus) in The First Workshop on the Algorithmic Foundations of
Robotics, A. K. Peters, Boston, MA. ed. R. Wilson and J.-C.Latombe (1994).

 Automatic Sensor
Configuration for Task-Directed Planning (with Amy Briggs), Proceedings 1994
IEEE International Conference on Robotics and Automation, San Diego, CA
(May 1994).
Automatic Sensor
Configuration for Task-Directed Planning (with Amy Briggs), Proceedings 1994
IEEE International Conference on Robotics and Automation, San Diego, CA
(May 1994).

 Mobile
Robot Self-Localization without Explicit Landmarks, (with R. G. Brown),
Algorithmica 26(3/4), March/April (2000) pp.
515-559.
Mobile
Robot Self-Localization without Explicit Landmarks, (with R. G. Brown),
Algorithmica 26(3/4), March/April (2000) pp.
515-559.

 Visibility-Based
Planning of Sensor Control Strategies, (with Amy Briggs),
Algorithmica 26(3/4), March/April (2000) pp.
364-388.
Visibility-Based
Planning of Sensor Control Strategies, (with Amy Briggs),
Algorithmica 26(3/4), March/April (2000) pp.
364-388.

 The Area Bisectors
of a Polygon, (with K.-F.
Böhringer, and D. Halperin) Second CGC Workshop on Computational Geometry,
(October 18-19, 1997) Duke University, Durham, NC.
The Area Bisectors
of a Polygon, (with K.-F.
Böhringer, and D. Halperin) Second CGC Workshop on Computational Geometry,
(October 18-19, 1997) Duke University, Durham, NC.

 K.-F.
Böhringer, B. R. Donald, and N. C. MacDonald, Upper and Lower
Bounds for Programmable Vector Fields with Applications to MEMS and Vibratory
Parts Feeders, [TR FTP]
International Workshop on the Algorithmic Foundations of Robotics,
Toulouse, France (1996). in Algorithms for Robotic Motion and
Manipulation,, ed. J. P. Laumond and M. Overmars, A. K. Peters, Wellesley,
MA (1997). pp. 255-276.
K.-F.
Böhringer, B. R. Donald, and N. C. MacDonald, Upper and Lower
Bounds for Programmable Vector Fields with Applications to MEMS and Vibratory
Parts Feeders, [TR FTP]
International Workshop on the Algorithmic Foundations of Robotics,
Toulouse, France (1996). in Algorithms for Robotic Motion and
Manipulation,, ed. J. P. Laumond and M. Overmars, A. K. Peters, Wellesley,
MA (1997). pp. 255-276.

 Amy Briggs and B. R. Donald, Robust Geometric
Algorithms for Sensor Planning, [TR FTP]
International Workshop on the Algorithmic Foundations of Robotics,
Toulouse, France (1996).
Amy Briggs and B. R. Donald, Robust Geometric
Algorithms for Sensor Planning, [TR FTP]
International Workshop on the Algorithmic Foundations of Robotics,
Toulouse, France (1996).

 Automatic Sensor
Configuration for Task-Directed Planning (with Amy Briggs), Proceedings 1994
IEEE International Conference on Robotics and Automation, San Diego, CA
(May 1994).
Automatic Sensor
Configuration for Task-Directed Planning (with Amy Briggs), Proceedings 1994
IEEE International Conference on Robotics and Automation, San Diego, CA
(May 1994).

 Part
Orientation with One or Two Stable Equilibria Using Programmable Force
Fields, (with K.-F.
Böhringer, F. Lamiraux, and L. Kavraki), IEEE Transactions on Robotics and
Automation, 16(2), April 2000, pp. 157-170.
Part
Orientation with One or Two Stable Equilibria Using Programmable Force
Fields, (with K.-F.
Böhringer, F. Lamiraux, and L. Kavraki), IEEE Transactions on Robotics and
Automation, 16(2), April 2000, pp. 157-170.

 Fully
Programmable MEMS Ciliary Actuator Arrays for Micromanipulation Tasks,
(with J. Suh, R. B. Darling, K.-F. Böhringer, H. Baltes, and G.Kovacs), the
IEEE International Conference on Robotics and Automation (ICRA) San
Francisco (April, 2000) pp. 1101-1108.
Fully
Programmable MEMS Ciliary Actuator Arrays for Micromanipulation Tasks,
(with J. Suh, R. B. Darling, K.-F. Böhringer, H. Baltes, and G.Kovacs), the
IEEE International Conference on Robotics and Automation (ICRA) San
Francisco (April, 2000) pp. 1101-1108.


 Sensorless
Manipulation Algorithms Using a Vibrating Surface, (with K.-F.
Böhringer, V. Bhatt, and K. Goldberg)
Algorithmica 26(3/4), March/April (2000) pp.
389-429.
Sensorless
Manipulation Algorithms Using a Vibrating Surface, (with K.-F.
Böhringer, V. Bhatt, and K. Goldberg)
Algorithmica 26(3/4), March/April (2000) pp.
389-429.

 CMOS Integrated
Organic Ciliary Actuator Arrays for General-Purpose Micromanipulation
Tasks, (with J. Suh, R. B. Darling, K.-F. Böhringer, H. Baltes, and G.
Kovacs) Journal of Microelectromechanical Systems, vol. 8, No. 4
(Dec. 1999), pp. 483-496.
CMOS Integrated
Organic Ciliary Actuator Arrays for General-Purpose Micromanipulation
Tasks, (with J. Suh, R. B. Darling, K.-F. Böhringer, H. Baltes, and G.
Kovacs) Journal of Microelectromechanical Systems, vol. 8, No. 4
(Dec. 1999), pp. 483-496.

 "A Single Universal
Force Field Can Uniquely Pose Any Part Up To Symmetry," (with K.-F.
Böhringer, F. Lamiraux, and L. Kavraki) 9th International Symposium of
Robotics Research (ISRR) Snowbird, Utah October 9-12
(1999).
"A Single Universal
Force Field Can Uniquely Pose Any Part Up To Symmetry," (with K.-F.
Böhringer, F. Lamiraux, and L. Kavraki) 9th International Symposium of
Robotics Research (ISRR) Snowbird, Utah October 9-12
(1999).

 We have three chapters in Distributed
Manipulation, ed. K. Böhringer
et al., Kluwer Academic Publishing, Boston (2000):
We have three chapters in Distributed
Manipulation, ed. K. Böhringer
et al., Kluwer Academic Publishing, Boston (2000):

 CMOS
Integrated Organic Ciliary Actuator Array as a General-Purpose
Micromanipulation Tool (with J. Suh, B. Darling, K.-F.
Böhringer, H. Baltes, and G. Kovacs) IEEE
International Conference on Robotics and Automation, Workshop on Distributed
Manipulation, (Detroit) May 11, 1999.
CMOS
Integrated Organic Ciliary Actuator Array as a General-Purpose
Micromanipulation Tool (with J. Suh, B. Darling, K.-F.
Böhringer, H. Baltes, and G. Kovacs) IEEE
International Conference on Robotics and Automation, Workshop on Distributed
Manipulation, (Detroit) May 11, 1999.

 Part
Orientation with One or Two Stable Equilibria Using Programmable Force
Fields, (with K.-F.
Böhringer, F. Lamiraux, and L. Kavraki), IEEE
International Conference on Robotics and Automation, Workshop on Distributed
Manipulation (Detroit) May 11, 1999.
Part
Orientation with One or Two Stable Equilibria Using Programmable Force
Fields, (with K.-F.
Böhringer, F. Lamiraux, and L. Kavraki), IEEE
International Conference on Robotics and Automation, Workshop on Distributed
Manipulation (Detroit) May 11, 1999.

 Algorithmic
MEMS, (with K.-F.
Böhringer), Invited lecture at the Third International
Workshop on the Algorithmic Foundations of Robotics (WAFR), Houston, TX
(March, 1998). Book version: In Robotics: The Algorithmic
Perspective, ed. P. Agarwal et al., A. K. Peters, Natick, MA
(1998). pp. 1-20.
Algorithmic
MEMS, (with K.-F.
Böhringer), Invited lecture at the Third International
Workshop on the Algorithmic Foundations of Robotics (WAFR), Houston, TX
(March, 1998). Book version: In Robotics: The Algorithmic
Perspective, ed. P. Agarwal et al., A. K. Peters, Natick, MA
(1998). pp. 1-20.

 Computational
Methods for the Design and Control of MEMS Micromanipulator Arrays (with K.-F.
Böhringer, G. Kovacs, N. MacDonald, and J. Suh) IEEE Computational
Science and Engineering, Vol. 4, No. 1. Special Issue on Computational
MEMS (Jan-March 1997). pp. 17-29.
Computational
Methods for the Design and Control of MEMS Micromanipulator Arrays (with K.-F.
Böhringer, G. Kovacs, N. MacDonald, and J. Suh) IEEE Computational
Science and Engineering, Vol. 4, No. 1. Special Issue on Computational
MEMS (Jan-March 1997). pp. 17-29.

 On The
Area Bisectors of a Polygon, (with K.-F.
Böhringer, and D. Halperin) Discrete and Computational Geometry, vol. 22
(1999), pp. 269-285.
On The
Area Bisectors of a Polygon, (with K.-F.
Böhringer, and D. Halperin) Discrete and Computational Geometry, vol. 22
(1999), pp. 269-285.

 Programmable
Vector Fields for Distributed Manipulation, with Applications to MEMS Actuator
Arrays and Vibratory Parts Feeders, [TR
FTP] (with K.-F.
Böhringer, and N. C. MacDonald), (International Journal of Robotics
Research). vol. 18(2), pp. 168-200 (1999).
Programmable
Vector Fields for Distributed Manipulation, with Applications to MEMS Actuator
Arrays and Vibratory Parts Feeders, [TR
FTP] (with K.-F.
Böhringer, and N. C. MacDonald), (International Journal of Robotics
Research). vol. 18(2), pp. 168-200 (1999).

 Micro Contacts and
Micro Manipulation with MEMS Actuator Arrays, (with K.-F.
Böhringer IEEE International Conference on Robotics and Automation,
Workshop on Modeling, Contact Analysis, and Simulation of Mechanical Systems
in Robotics and Manufacturing, (Belgium, 1998).
Micro Contacts and
Micro Manipulation with MEMS Actuator Arrays, (with K.-F.
Böhringer IEEE International Conference on Robotics and Automation,
Workshop on Modeling, Contact Analysis, and Simulation of Mechanical Systems
in Robotics and Manufacturing, (Belgium, 1998).

 The Area Bisectors
of a Polygon, (with K.-F.
Böhringer, and D. Halperin) Second CGC Workshop on Computational Geometry,
(October 18-19, 1997) Duke University, Durham, NC.
The Area Bisectors
of a Polygon, (with K.-F.
Böhringer, and D. Halperin) Second CGC Workshop on Computational Geometry,
(October 18-19, 1997) Duke University, Durham, NC.

 Vector
Fields for Task-Level Distributed Manipulation: Experiments with Organic Micro
Actuator Arrays [TR
FTP], (with K.-F.
Böhringer, J. Suh, and G. Kovacs) in Proc. IEEE International Conference
on Robotics and Automation, Albuquerque, NM (1997). pp 1779-1786.
Vector
Fields for Task-Level Distributed Manipulation: Experiments with Organic Micro
Actuator Arrays [TR
FTP], (with K.-F.
Böhringer, J. Suh, and G. Kovacs) in Proc. IEEE International Conference
on Robotics and Automation, Albuquerque, NM (1997). pp 1779-1786.

 The Area
Bisectors of a Polygon and Force Equilibria in Programmable Vector Fields
[TR
FTP], (with K.-F.
Böhringer, and D. Halperin) 13th Proc. ACM Symposium on Computational
Geometry, Nice, France (1997) pp. 457-459.
The Area
Bisectors of a Polygon and Force Equilibria in Programmable Vector Fields
[TR
FTP], (with K.-F.
Böhringer, and D. Halperin) 13th Proc. ACM Symposium on Computational
Geometry, Nice, France (1997) pp. 457-459.

 K.-F.
Böhringer, B. R. Donald, and N. C. MacDonald, Upper and Lower
Bounds for Programmable Vector Fields with Applications to MEMS and Vibratory
Parts Feeders, [TR FTP]
International Workshop on the Algorithmic Foundations of Robotics,
Toulouse, France (1996). in Algorithms for Robotic Motion and
Manipulation,, ed. J. P. Laumond and M. Overmars, A. K. Peters, Wellesley,
MA (1997). pp. 255-276.
K.-F.
Böhringer, B. R. Donald, and N. C. MacDonald, Upper and Lower
Bounds for Programmable Vector Fields with Applications to MEMS and Vibratory
Parts Feeders, [TR FTP]
International Workshop on the Algorithmic Foundations of Robotics,
Toulouse, France (1996). in Algorithms for Robotic Motion and
Manipulation,, ed. J. P. Laumond and M. Overmars, A. K. Peters, Wellesley,
MA (1997). pp. 255-276.

 K.-F.
Böhringer, B. R. Donald, and N. C. MacDonald, Single-Crystal Silicon
Actuator Arrays for Micro Manipulation Tasks [TR FTP], IEEE
Workshop on Micro Electro Mechanical Systems (MEMS), San Diego, California
(February 1996).
K.-F.
Böhringer, B. R. Donald, and N. C. MacDonald, Single-Crystal Silicon
Actuator Arrays for Micro Manipulation Tasks [TR FTP], IEEE
Workshop on Micro Electro Mechanical Systems (MEMS), San Diego, California
(February 1996).
 Structure of Dihydrofolate Reductase-Thymidylate Synthase
from Cryptosporidium hominis Reveals a Novel Architecture for the
Bifunctional Enzyme, (with R. O'Neil, R. Lilien, R. Stroud, and A. Anderson)
Journal of Eukaryotic Microbiology 2003;
50(6):555-556. Cover
article.
Structure of Dihydrofolate Reductase-Thymidylate Synthase
from Cryptosporidium hominis Reveals a Novel Architecture for the
Bifunctional Enzyme, (with R. O'Neil, R. Lilien, R. Stroud, and A. Anderson)
Journal of Eukaryotic Microbiology 2003;
50(6):555-556. Cover
article.




|
(in Comm. ACM, 2019). |
This journal paper is a revised paper based on the earlier Conference version:
LUTE (Local Unpruned Tuple Expansion): Accurate continuously flexible protein design with general energy functions and rigid-rotamer-like efficiency (with M. Hallen and J. Jou). Proceedings of the Annual International Conference on Research in Computational Molecular Biology (RECOMB), Los Angeles, CA, April, 2016. In Research in Computational Molecular Biology, Lecture Notes in Bioinformatics (LNBI), Springer-Verlag (Berlin), Volume 9649, 2016, pp. 122-136.
This journal paper is a revised paper based on the earlier Conference version:
COMETS (Constrained Optimization of Multistate Energies by Tree Search): A provable and efficient algorithm to optimize binding affinity and specificity with respect to sequence (with M. Hallen). Proceedings of the Annual International Conference on Research in Computational Molecular Biology (RECOMB), Warsaw, April 12-15, 2015. In Research in Computational Molecular Biology Lecture Notes in Computer Science, Springer-Verlag (Berlin), Volume 9029, 2015, pp 122-135.
This journal paper is a revised paper based on the earlier Conference version:
BWM*: A Novel, Provable, Ensemble-based Dynamic Programming Algorithm for Sparse Approximations of Computational Protein Design (with J. Jou, S. Jain, and I. Georgiev). Proceedings of the Annual International Conference on Research in Computational Molecular Biology (RECOMB), Warsaw, April 12-15, 2015. In Research in Computational Molecular Biology Lecture Notes in Computer Science, Springer-Verlag (Berlin), Volume 9029, 2015, pp 154-166.
 |
||
| Nuclear Vector
Replacement. |
||
Scientific papers (with some pointers to videos) are provided for you here, in the list immediately below:

 Part
Orientation with One or Two Stable Equilibria Using Programmable Force
Fields, (with K.-F.
Böhringer, F. Lamiraux, and L. Kavraki), IEEE Transactions on Robotics and
Automation, 16(2), April 2000, pp. 157-170.
Part
Orientation with One or Two Stable Equilibria Using Programmable Force
Fields, (with K.-F.
Böhringer, F. Lamiraux, and L. Kavraki), IEEE Transactions on Robotics and
Automation, 16(2), April 2000, pp. 157-170.

 Fully
Programmable MEMS Ciliary Actuator Arrays for Micromanipulation Tasks,
(with J. Suh, R. B. Darling, K.-F. Böhringer, H. Baltes, and G.Kovacs), the
IEEE International Conference on Robotics and Automation (ICRA) San
Francisco (April, 2000) pp. 1101-1108.
Fully
Programmable MEMS Ciliary Actuator Arrays for Micromanipulation Tasks,
(with J. Suh, R. B. Darling, K.-F. Böhringer, H. Baltes, and G.Kovacs), the
IEEE International Conference on Robotics and Automation (ICRA) San
Francisco (April, 2000) pp. 1101-1108.


 Sensorless
Manipulation Algorithms Using a Vibrating Surface, (with K.-F.
Böhringer, V. Bhatt, and K. Goldberg)
Algorithmica 26(3/4), March/April (2000) pp.
389-429.
Sensorless
Manipulation Algorithms Using a Vibrating Surface, (with K.-F.
Böhringer, V. Bhatt, and K. Goldberg)
Algorithmica 26(3/4), March/April (2000) pp.
389-429.

 CMOS Integrated
Organic Ciliary Actuator Arrays for General-Purpose Micromanipulation
Tasks, (with J. Suh, R. B. Darling, K.-F. Böhringer, H. Baltes, and G.
Kovacs) Journal of Microelectromechanical Systems, vol. 8, No. 4
(Dec. 1999), pp. 483-496.
CMOS Integrated
Organic Ciliary Actuator Arrays for General-Purpose Micromanipulation
Tasks, (with J. Suh, R. B. Darling, K.-F. Böhringer, H. Baltes, and G.
Kovacs) Journal of Microelectromechanical Systems, vol. 8, No. 4
(Dec. 1999), pp. 483-496.

 "A Single Universal
Force Field Can Uniquely Pose Any Part Up To Symmetry," (with K.-F.
Böhringer, F. Lamiraux, and L. Kavraki) 9th International Symposium of
Robotics Research (ISRR) Snowbird, Utah October 9-12
(1999).
"A Single Universal
Force Field Can Uniquely Pose Any Part Up To Symmetry," (with K.-F.
Böhringer, F. Lamiraux, and L. Kavraki) 9th International Symposium of
Robotics Research (ISRR) Snowbird, Utah October 9-12
(1999).

 We have three chapters in Distributed
Manipulation, ed. K. Böhringer
et al., Kluwer Academic Publishing, Boston (2000):
We have three chapters in Distributed
Manipulation, ed. K. Böhringer
et al., Kluwer Academic Publishing, Boston (2000):

 CMOS
Integrated Organic Ciliary Actuator Array as a General-Purpose
Micromanipulation Tool (with J. Suh, B. Darling, K.-F.
Böhringer, H. Baltes, and G. Kovacs) IEEE
International Conference on Robotics and Automation, Workshop on Distributed
Manipulation, (Detroit) May 11, 1999.
CMOS
Integrated Organic Ciliary Actuator Array as a General-Purpose
Micromanipulation Tool (with J. Suh, B. Darling, K.-F.
Böhringer, H. Baltes, and G. Kovacs) IEEE
International Conference on Robotics and Automation, Workshop on Distributed
Manipulation, (Detroit) May 11, 1999.

 Part
Orientation with One or Two Stable Equilibria Using Programmable Force
Fields, (with K.-F.
Böhringer, F. Lamiraux, and L. Kavraki), IEEE
International Conference on Robotics and Automation, Workshop on Distributed
Manipulation (Detroit) May 11, 1999.
Part
Orientation with One or Two Stable Equilibria Using Programmable Force
Fields, (with K.-F.
Böhringer, F. Lamiraux, and L. Kavraki), IEEE
International Conference on Robotics and Automation, Workshop on Distributed
Manipulation (Detroit) May 11, 1999.

 Algorithmic
MEMS, (with K.-F.
Böhringer), Invited lecture at the Third International
Workshop on the Algorithmic Foundations of Robotics (WAFR), Houston, TX
(March, 1998). Book version: In Robotics: The Algorithmic
Perspective, ed. P. Agarwal et al., A. K. Peters, Natick, MA
(1998). pp. 1-20.
Algorithmic
MEMS, (with K.-F.
Böhringer), Invited lecture at the Third International
Workshop on the Algorithmic Foundations of Robotics (WAFR), Houston, TX
(March, 1998). Book version: In Robotics: The Algorithmic
Perspective, ed. P. Agarwal et al., A. K. Peters, Natick, MA
(1998). pp. 1-20.

 Computational
Methods for the Design and Control of MEMS Micromanipulator Arrays (with K.-F.
Böhringer, G. Kovacs, N. MacDonald, and J. Suh) IEEE Computational
Science and Engineering, Vol. 4, No. 1. Special Issue on Computational
MEMS (Jan-March 1997). pp. 17-29.
Computational
Methods for the Design and Control of MEMS Micromanipulator Arrays (with K.-F.
Böhringer, G. Kovacs, N. MacDonald, and J. Suh) IEEE Computational
Science and Engineering, Vol. 4, No. 1. Special Issue on Computational
MEMS (Jan-March 1997). pp. 17-29.

 On The
Area Bisectors of a Polygon, (with K.-F.
Böhringer, and D. Halperin) Discrete and Computational Geometry, vol. 22
(1999), pp. 269-285.
On The
Area Bisectors of a Polygon, (with K.-F.
Böhringer, and D. Halperin) Discrete and Computational Geometry, vol. 22
(1999), pp. 269-285.

 Programmable
Vector Fields for Distributed Manipulation, with Applications to MEMS Actuator
Arrays and Vibratory Parts Feeders, [TR
FTP] (with K.-F.
Böhringer, and N. C. MacDonald), (International Journal of Robotics
Research). vol. 18(2), pp. 168-200 (1999).
Programmable
Vector Fields for Distributed Manipulation, with Applications to MEMS Actuator
Arrays and Vibratory Parts Feeders, [TR
FTP] (with K.-F.
Böhringer, and N. C. MacDonald), (International Journal of Robotics
Research). vol. 18(2), pp. 168-200 (1999).

 Micro Contacts and
Micro Manipulation with MEMS Actuator Arrays, (with K.-F.
Böhringer IEEE International Conference on Robotics and Automation,
Workshop on Modeling, Contact Analysis, and Simulation of Mechanical Systems
in Robotics and Manufacturing, (Belgium, 1998).
Micro Contacts and
Micro Manipulation with MEMS Actuator Arrays, (with K.-F.
Böhringer IEEE International Conference on Robotics and Automation,
Workshop on Modeling, Contact Analysis, and Simulation of Mechanical Systems
in Robotics and Manufacturing, (Belgium, 1998).

 The Area Bisectors
of a Polygon, (with K.-F.
Böhringer, and D. Halperin) Second CGC Workshop on Computational Geometry,
(October 18-19, 1997) Duke University, Durham, NC.
The Area Bisectors
of a Polygon, (with K.-F.
Böhringer, and D. Halperin) Second CGC Workshop on Computational Geometry,
(October 18-19, 1997) Duke University, Durham, NC.

 Vector
Fields for Task-Level Distributed Manipulation: Experiments with Organic Micro
Actuator Arrays [TR
FTP], (with K.-F.
Böhringer, J. Suh, and G. Kovacs) in Proc. IEEE International Conference
on Robotics and Automation, Albuquerque, NM (1997). pp 1779-1786.
Vector
Fields for Task-Level Distributed Manipulation: Experiments with Organic Micro
Actuator Arrays [TR
FTP], (with K.-F.
Böhringer, J. Suh, and G. Kovacs) in Proc. IEEE International Conference
on Robotics and Automation, Albuquerque, NM (1997). pp 1779-1786.

 The Area
Bisectors of a Polygon and Force Equilibria in Programmable Vector Fields
[TR
FTP], (with K.-F.
Böhringer, and D. Halperin) 13th Proc. ACM Symposium on Computational
Geometry, Nice, France (1997) pp. 457-459.
The Area
Bisectors of a Polygon and Force Equilibria in Programmable Vector Fields
[TR
FTP], (with K.-F.
Böhringer, and D. Halperin) 13th Proc. ACM Symposium on Computational
Geometry, Nice, France (1997) pp. 457-459.

 K.-F.
Böhringer, B. R. Donald, and N. C. MacDonald, Upper and Lower
Bounds for Programmable Vector Fields with Applications to MEMS and Vibratory
Parts Feeders, [TR FTP]
International Workshop on the Algorithmic Foundations of Robotics,
Toulouse, France (1996). in Algorithms for Robotic Motion and
Manipulation,, ed. J. P. Laumond and M. Overmars, A. K. Peters, Wellesley,
MA (1997). pp. 255-276.
K.-F.
Böhringer, B. R. Donald, and N. C. MacDonald, Upper and Lower
Bounds for Programmable Vector Fields with Applications to MEMS and Vibratory
Parts Feeders, [TR FTP]
International Workshop on the Algorithmic Foundations of Robotics,
Toulouse, France (1996). in Algorithms for Robotic Motion and
Manipulation,, ed. J. P. Laumond and M. Overmars, A. K. Peters, Wellesley,
MA (1997). pp. 255-276.

 K.-F.
Böhringer, B. R. Donald, and N. C. MacDonald, Single-Crystal Silicon
Actuator Arrays for Micro Manipulation Tasks [TR FTP], IEEE
Workshop on Micro Electro Mechanical Systems (MEMS), San Diego, California
(February 1996).
K.-F.
Böhringer, B. R. Donald, and N. C. MacDonald, Single-Crystal Silicon
Actuator Arrays for Micro Manipulation Tasks [TR FTP], IEEE
Workshop on Micro Electro Mechanical Systems (MEMS), San Diego, California
(February 1996).

 K.-F.
Böhringer, B. R. Donald, and N. C. MacDonald, Classification
and Lower Bounds for MEMS Arrays and Vibratory Parts Feeders: What
Programmable Vector Fields Can (and Cannot) Do, IEEE
International Conference on Robotics and Automation (ICRA), Minneapolis,
Minnesota (April 1996).
K.-F.
Böhringer, B. R. Donald, and N. C. MacDonald, Classification
and Lower Bounds for MEMS Arrays and Vibratory Parts Feeders: What
Programmable Vector Fields Can (and Cannot) Do, IEEE
International Conference on Robotics and Automation (ICRA), Minneapolis,
Minnesota (April 1996).


 Distributed Robotic
Manipulation: Experiments in Minimalism [TR FTP], in
International Symposium on Experimental Robotics, (ISER) Stanford, CA
(1995). Appears in: in Experimental Robotics IV, Lecture Notes in Control
and Information Sciences 223, ed. O. Khatib et al., Springer Verlag
(Berlin) 1997 pp. 11-25.
Distributed Robotic
Manipulation: Experiments in Minimalism [TR FTP], in
International Symposium on Experimental Robotics, (ISER) Stanford, CA
(1995). Appears in: in Experimental Robotics IV, Lecture Notes in Control
and Information Sciences 223, ed. O. Khatib et al., Springer Verlag
(Berlin) 1997 pp. 11-25.

 Sensorless Manipulation Using Massively Parallel Micro-fabricated Actuator
Arrays [TR
FTP] (with K.-F.
Böhringer, R. Mihailovich, and Noel C. MacDonald), Proc. IEEE
International Conference on Robotics and Automation, San Diego, CA (May,
1994).
Sensorless Manipulation Using Massively Parallel Micro-fabricated Actuator
Arrays [TR
FTP] (with K.-F.
Böhringer, R. Mihailovich, and Noel C. MacDonald), Proc. IEEE
International Conference on Robotics and Automation, San Diego, CA (May,
1994). Scientific papers (with some pointers to videos) are provided for you here, in the list immediately below:
 Distributed
Manipulation of Multiple Objects Using Ropes, (with L. Gariepy and D.
Rus), Proc. IEEE International Conference on Robotics and Automation
(ICRA), San Francisco (April, 2000) pp. 450-457.
Distributed
Manipulation of Multiple Objects Using Ropes, (with L. Gariepy and D.
Rus), Proc. IEEE International Conference on Robotics and Automation
(ICRA), San Francisco (April, 2000) pp. 450-457.
| Movie, 4:30, Quicktime w/Sorenson codec, max 250 kb/s (42MB) |
mapmaking-movie-5.250kbit.mov |
| Movie, 4:30, Quicktime w/Sorenson codec, max 100 kb/s (26MB) |
mapmaking-movie-5.100kbit.mov |
| The paper (Gzipped PostScript) (0.8 MB) | ICRA'2000 paper |

 Visibility-Based
Planning of Sensor Control Strategies, (with Amy Briggs),
Algorithmica 26(3/4), March/April (2000) pp.
364-388.
Visibility-Based
Planning of Sensor Control Strategies, (with Amy Briggs),
Algorithmica 26(3/4), March/April (2000) pp.
364-388.

 Mobile
Robot Self-Localization without Explicit Landmarks, (with R. G. Brown),
Algorithmica 26(3/4), March/April (2000) pp.
515-559.
Mobile
Robot Self-Localization without Explicit Landmarks, (with R. G. Brown),
Algorithmica 26(3/4), March/April (2000) pp.
515-559.
 Experiments in
Constrained Prehensile Manipulation: Distributed Manipulation with Ropes,
with Larry Gariepy and Daniela
Rus, In Experimental Robotics VI, Lecture Notes in Control and Information
Sciences, Vol. 250, eds. P. Corke, Springer-Verlag, (2000).
Experiments in
Constrained Prehensile Manipulation: Distributed Manipulation with Ropes,
with Larry Gariepy and Daniela
Rus, In Experimental Robotics VI, Lecture Notes in Control and Information
Sciences, Vol. 250, eds. P. Corke, Springer-Verlag, (2000).

 Amy Briggs and B. R. Donald, Robust Geometric
Algorithms for Sensor Planning, [TR FTP]
International Workshop on the Algorithmic Foundations of Robotics,
Toulouse, France (1996).
Amy Briggs and B. R. Donald, Robust Geometric
Algorithms for Sensor Planning, [TR FTP]
International Workshop on the Algorithmic Foundations of Robotics,
Toulouse, France (1996).
 Information
Invariants for Distributed Manipulation (with J. Jennings and D. Rus) in International
Journal of Robotics Research, Vol. 16, No. 5 (1997). pp. 673-702.
Information
Invariants for Distributed Manipulation (with J. Jennings and D. Rus) in International
Journal of Robotics Research, Vol. 16, No. 5 (1997). pp. 673-702.
 B. R. Donald, J. Jennings, and D. Rus, Minimalism +
Distribution = Supermodularity [TR FTP], Journal
of Experimental and Theoretical Artificial Intelligence (JETAI), Special
issue on Software Architectures for Hardware Agents; Vol. 9 No. 2-3 (1997) pp.
293-321.
B. R. Donald, J. Jennings, and D. Rus, Minimalism +
Distribution = Supermodularity [TR FTP], Journal
of Experimental and Theoretical Artificial Intelligence (JETAI), Special
issue on Software Architectures for Hardware Agents; Vol. 9 No. 2-3 (1997) pp.
293-321.
 Cost-effective, Capable, and
Configurable Multiple Robots: A Report to the ARPA ISAT Study
Group, Summer, 1996 (with J. Jennings and D. Rus).
Cost-effective, Capable, and
Configurable Multiple Robots: A Report to the ARPA ISAT Study
Group, Summer, 1996 (with J. Jennings and D. Rus).
 Moving Furniture with
Teams of Automonous Mobile Robots [TR FTP], (with J.
Jennings and D. Rus) in
Moving Furniture with
Teams of Automonous Mobile Robots [TR FTP], (with J.
Jennings and D. Rus) in
Proc. IEEE/Robotics Society of Japan International Workshop on Intelligent Robots and Systems, (IROS) Pittsburgh, PA (1995).


 Information
Invariants for Distributed Manipulation [TR
FTP] (with J. Jennings and D.
Rus) in The First Workshop on the Algorithmic Foundations of
Robotics, A. K. Peters, Boston, MA. ed. R. Wilson and J.-C.Latombe (1994).
Information
Invariants for Distributed Manipulation [TR
FTP] (with J. Jennings and D.
Rus) in The First Workshop on the Algorithmic Foundations of
Robotics, A. K. Peters, Boston, MA. ed. R. Wilson and J.-C.Latombe (1994).

 Automatic Sensor
Configuration for Task-Directed Planning (with Amy Briggs), Proceedings 1994
IEEE International Conference on Robotics and Automation, San Diego, CA
(May 1994).
Automatic Sensor
Configuration for Task-Directed Planning (with Amy Briggs), Proceedings 1994
IEEE International Conference on Robotics and Automation, San Diego, CA
(May 1994).

 Mobile
Robot Self-Localization without Explicit Landmarks, (with R. G. Brown),
Algorithmica 26(3/4), March/April (2000) pp.
515-559.
Mobile
Robot Self-Localization without Explicit Landmarks, (with R. G. Brown),
Algorithmica 26(3/4), March/April (2000) pp.
515-559.

 Visibility-Based
Planning of Sensor Control Strategies, (with Amy Briggs),
Algorithmica 26(3/4), March/April (2000) pp.
364-388.
Visibility-Based
Planning of Sensor Control Strategies, (with Amy Briggs),
Algorithmica 26(3/4), March/April (2000) pp.
364-388.

 The Area Bisectors
of a Polygon, (with K.-F.
Böhringer, and D. Halperin) Second CGC Workshop on Computational Geometry,
(October 18-19, 1997) Duke University, Durham, NC.
The Area Bisectors
of a Polygon, (with K.-F.
Böhringer, and D. Halperin) Second CGC Workshop on Computational Geometry,
(October 18-19, 1997) Duke University, Durham, NC.

 K.-F.
Böhringer, B. R. Donald, and N. C. MacDonald, Upper and Lower
Bounds for Programmable Vector Fields with Applications to MEMS and Vibratory
Parts Feeders, [TR FTP]
International Workshop on the Algorithmic Foundations of Robotics,
Toulouse, France (1996). in Algorithms for Robotic Motion and
Manipulation,, ed. J. P. Laumond and M. Overmars, A. K. Peters, Wellesley,
MA (1997). pp. 255-276.
K.-F.
Böhringer, B. R. Donald, and N. C. MacDonald, Upper and Lower
Bounds for Programmable Vector Fields with Applications to MEMS and Vibratory
Parts Feeders, [TR FTP]
International Workshop on the Algorithmic Foundations of Robotics,
Toulouse, France (1996). in Algorithms for Robotic Motion and
Manipulation,, ed. J. P. Laumond and M. Overmars, A. K. Peters, Wellesley,
MA (1997). pp. 255-276.

 Amy Briggs and B. R. Donald, Robust Geometric
Algorithms for Sensor Planning, [TR FTP]
International Workshop on the Algorithmic Foundations of Robotics,
Toulouse, France (1996).
Amy Briggs and B. R. Donald, Robust Geometric
Algorithms for Sensor Planning, [TR FTP]
International Workshop on the Algorithmic Foundations of Robotics,
Toulouse, France (1996).
Here are 3 of our papers on Kinodynamic Planning:

 Automatic Sensor
Configuration for Task-Directed Planning (with Amy Briggs), Proceedings 1994
IEEE International Conference on Robotics and Automation, San Diego, CA
(May 1994).
Automatic Sensor
Configuration for Task-Directed Planning (with Amy Briggs), Proceedings 1994
IEEE International Conference on Robotics and Automation, San Diego, CA
(May 1994).
Scientific papers (with some pointers to videos) are provided for you here, in the list immediately below:

 "Accessible
Animation and Customizable Graphics via Simplicial Configuration Modeling"
(with T. Ngo, D. Cutrell, J. Dana, L. Loeb, and S. Zhu), in Proc. ACM
SIGGRAPH (New Orleans) July, 2000, pp. 403-410.
"Accessible
Animation and Customizable Graphics via Simplicial Configuration Modeling"
(with T. Ngo, D. Cutrell, J. Dana, L. Loeb, and S. Zhu), in Proc. ACM
SIGGRAPH (New Orleans) July, 2000, pp. 403-410.
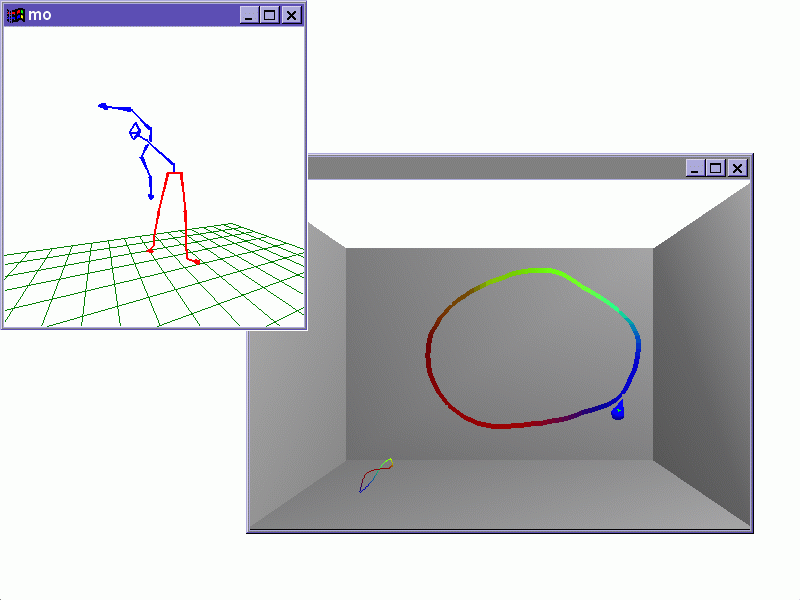


 Using Haptic
Vector Fields for Animation Motion Control (with F. Henle) Proc. IEEE
Int. Conf. on Robotics and Automation (ICRA), San Francisco (April, 2000)
pp. 3435-3442.
Using Haptic
Vector Fields for Animation Motion Control (with F. Henle) Proc. IEEE
Int. Conf. on Robotics and Automation (ICRA), San Francisco (April, 2000)
pp. 3435-3442.

 Prototyping
Animation Motion Control with Haptic Programmable Force Fields, Workshop
on "Motion Support in Virtual Prototyping," (with Fred Henle) Stanford, May 5-7,
1999.
Prototyping
Animation Motion Control with Haptic Programmable Force Fields, Workshop
on "Motion Support in Virtual Prototyping," (with Fred Henle) Stanford, May 5-7,
1999.

 System for Image
Manipulation and Animation Using Embedded Constraint Graphics (with J. T.
Ngo), U.S. Patent #5,933,150, issued August 3, 1999.
System for Image
Manipulation and Animation Using Embedded Constraint Graphics (with J. T.
Ngo), U.S. Patent #5,933,150, issued August 3, 1999.


 Mobile
Robot Self-Localization without Explicit Landmarks, (with R. G. Brown),
Algorithmica 26(3/4), March/April (2000) pp.
515-559.
Mobile
Robot Self-Localization without Explicit Landmarks, (with R. G. Brown),
Algorithmica 26(3/4), March/April (2000) pp.
515-559.

 Visibility-Based
Planning of Sensor Control Strategies, (with Amy Briggs),
Algorithmica 26(3/4), March/April (2000) pp.
364-388.
Visibility-Based
Planning of Sensor Control Strategies, (with Amy Briggs),
Algorithmica 26(3/4), March/April (2000) pp.
364-388.

 Part
Orientation with One or Two Stable Equilibria Using Programmable Force
Fields, (with K.-F.
Böhringer, F. Lamiraux, and L. Kavraki), IEEE Transactions on Robotics and
Automation, 16(2), April 2000, pp. 157-170.
Part
Orientation with One or Two Stable Equilibria Using Programmable Force
Fields, (with K.-F.
Böhringer, F. Lamiraux, and L. Kavraki), IEEE Transactions on Robotics and
Automation, 16(2), April 2000, pp. 157-170.

 "A Single Universal
Force Field Can Uniquely Pose Any Part Up To Symmetry," (with K.-F.
Böhringer, F. Lamiraux, and L. Kavraki) 9th International Symposium of
Robotics Research (ISRR) Snowbird, Utah October 9-12 (1999).
"A Single Universal
Force Field Can Uniquely Pose Any Part Up To Symmetry," (with K.-F.
Böhringer, F. Lamiraux, and L. Kavraki) 9th International Symposium of
Robotics Research (ISRR) Snowbird, Utah October 9-12 (1999).

 On The
Area Bisectors of a Polygon, (with K.-F.
Böhringer, and D. Halperin) Discrete and Computational Geometry, vol. 22
(1999), pp. 269-285.
On The
Area Bisectors of a Polygon, (with K.-F.
Böhringer, and D. Halperin) Discrete and Computational Geometry, vol. 22
(1999), pp. 269-285.

 Part Orientation with
One or Two Stable Equilibria , (with K.-F.
Böhringer, F. Lamiraux, and L. Kavraki), International Conference on
Robotics and Automation, Workshop on Distributed Manipulation (Detroit)
May 11, 1999.
Part Orientation with
One or Two Stable Equilibria , (with K.-F.
Böhringer, F. Lamiraux, and L. Kavraki), International Conference on
Robotics and Automation, Workshop on Distributed Manipulation (Detroit)
May 11, 1999.

 The Area Bisectors
of a Polygon, (with K.-F.
Böhringer, and D. Halperin) Second CGC Workshop on Computational Geometry,
(October 18-19, 1997) Duke University, Durham, NC.
The Area Bisectors
of a Polygon, (with K.-F.
Böhringer, and D. Halperin) Second CGC Workshop on Computational Geometry,
(October 18-19, 1997) Duke University, Durham, NC.

 The Area
Bisectors of a Polygon and Force Equilibria in Programmable Vector Fields
[TR
FTP], (with K.-F.
Böhringer, and D. Halperin) 13th Proc. ACM Symposium on Computational
Geometry, Nice, France (1997) pp. 457-459.
The Area
Bisectors of a Polygon and Force Equilibria in Programmable Vector Fields
[TR
FTP], (with K.-F.
Böhringer, and D. Halperin) 13th Proc. ACM Symposium on Computational
Geometry, Nice, France (1997) pp. 457-459.

 Programmable
Vector Fields for Distributed Manipulation, with Applications to MEMS Actuator
Arrays and Vibratory Parts Feeders, [TR
FTP] (with K.-F.
Böhringer, and N. C. MacDonald), (International Journal of Robotics
Research, 1999) vol. 18(2) pp. 168-200.
Programmable
Vector Fields for Distributed Manipulation, with Applications to MEMS Actuator
Arrays and Vibratory Parts Feeders, [TR
FTP] (with K.-F.
Böhringer, and N. C. MacDonald), (International Journal of Robotics
Research, 1999) vol. 18(2) pp. 168-200.

 K.-F.
Böhringer, B. R. Donald, and N. C. MacDonald, Upper and Lower
Bounds for Programmable Vector Fields with Applications to MEMS and Vibratory
Parts Feeders, [TR FTP]
International Workshop on the Algorithmic Foundations of Robotics,
Toulouse, France (1996). in Algorithms for Robotic Motion and
Manipulation,, ed. J. P. Laumond and M. Overmars, A. K. Peters, Wellesley,
MA (1997). pp. 255-276.
K.-F.
Böhringer, B. R. Donald, and N. C. MacDonald, Upper and Lower
Bounds for Programmable Vector Fields with Applications to MEMS and Vibratory
Parts Feeders, [TR FTP]
International Workshop on the Algorithmic Foundations of Robotics,
Toulouse, France (1996). in Algorithms for Robotic Motion and
Manipulation,, ed. J. P. Laumond and M. Overmars, A. K. Peters, Wellesley,
MA (1997). pp. 255-276.

 Amy Briggs and B. R. Donald, Robust Geometric
Algorithms for Sensor Planning, [TR FTP]
International Workshop on the Algorithmic Foundations of Robotics,
Toulouse, France (1996).
Amy Briggs and B. R. Donald, Robust Geometric
Algorithms for Sensor Planning, [TR FTP]
International Workshop on the Algorithmic Foundations of Robotics,
Toulouse, France (1996).

 Automatic Sensor
Configuration for Task-Directed Planning (with Amy Briggs), Proceedings 1994
IEEE International Conference on Robotics and Automation, San Diego, CA
(May 1994).
Automatic Sensor
Configuration for Task-Directed Planning (with Amy Briggs), Proceedings 1994
IEEE International Conference on Robotics and Automation, San Diego, CA
(May 1994).
The lab YouTube channel http://www.youtube.com/user/labdonald/videos, has many videos describing our research, starting around 1988.
My C.V. (PDF) contains a list of publications, including a complete list those prior to 1994 (prior to 2015, actually).
Error Detection and Recovery in Robotics, Lecture Notes in
Computer Science, Vol. 336, Springer-Verlag, New York (1989). 314 pp.
ISBN
0387969098